Ex1.
!wget https://archive.ics.uci.edu/static/public/1/abalone.zip
!unzip abalone.zip
import pandas as pd
attr_names = (
"Sex", "Length(mm)", "Diameter(mm)", "Height(mm)", "Whole_weight(g)", "Shucked_weight(g)", "Viscera_weight(g)", "Shell_weight(g)", "Rings"
)
# Below shows the most commonly used parameters and kwargs of `pd.read_csv()`
data = pd.read_csv("abalone.data", sep=",", header=None, names=attr_names)
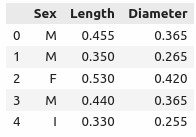
data
Output:
Sex Length(mm) Diameter(mm) Height(mm) Whole_weight(g) Shucked_weight(g) Viscera_weight(g) Shell_weight(g) Rings
0 M 0.455 0.365 0.095 0.5140 0.2245 0.1010 0.1500 15
1 M 0.350 0.265 0.090 0.2255 0.0995 0.0485 0.0700 7
2 F 0.530 0.420 0.135 0.6770 0.2565 0.1415 0.2100 9
3 M 0.440 0.365 0.125 0.5160 0.2155 0.1140 0.1550 10
4 I 0.330 0.255 0.080 0.2050 0.0895 0.0395 0.0550 7
... ... ... ... ... ... ... ... ... ...
4172 F 0.565 0.450 0.165 0.8870 0.3700 0.2390 0.2490 11
4173 M 0.590 0.440 0.135 0.9660 0.4390 0.2145 0.2605 10
4174 M 0.600 0.475 0.205 1.1760 0.5255 0.2875 0.3080 9
4175 F 0.625 0.485 0.150 1.0945 0.5310 0.2610 0.2960 10
4176 M 0.710 0.555 0.195 1.9485 0.9455 0.3765 0.4950 12
4177 rows × 9 columns
# Checking missing values in each column, a positive sum indicates the existence of missing entries in a given column
check_na = data.isna()
check_na.sum(axis=0)
Output:
Sex 0
Length(mm) 0
Diameter(mm) 0
Height(mm) 0
Whole_weight(g) 0
Shucked_weight(g) 0
Viscera_weight(g) 0
Shell_weight(g) 0
Rings 0
dtype: int64
- We don’t have any missing value in this abalone dataset.
- As per the introductory page of the dataset, there are totally 9 attributes (columns), with 8 of them numerical.
- The first column,
Sex, is categorical. We might explore all its possible values via the value_counts() method.
categories = data["Sex"].value_counts()
categories
Output:
M 1528
I 1342
F 1307
Name: Sex, dtype: int64
Ex2.
# access a column using dict-like indexing
data["Sex"][0], data.Rings[2]
Output:
('M', 9)
Ex4.
- Too much uniqueness of categorical values indicate that the amount of information that the feature carries is poor. We may safely exclude a feature if all its values are unique.
- Manual or automatic concatenation of categories might be required.
I thank @Tejas-Garhewal for his/her instructive post on this question.
Ex5.
Here is the cute cat.png file used. 

from PIL import Image
import numpy as np
img = Image.open("cat.png")
np.asarray(img)
Output:
array([[[199, 56, 130, 255],
[199, 56, 130, 255],
[199, 56, 130, 255],
...,
[202, 68, 130, 255],
[202, 68, 130, 255],
[202, 68, 130, 255]],
[[199, 56, 130, 255],
[199, 56, 130, 255],
[199, 56, 130, 255],
...,
[202, 68, 130, 255],
[202, 68, 130, 255],
[202, 68, 130, 255]],
[[199, 56, 130, 255],
[199, 56, 130, 255],
[199, 56, 130, 255],
...,
[202, 68, 130, 255],
[202, 68, 130, 255],
[202, 68, 130, 255]],
...,
[[233, 235, 236, 255],
[233, 235, 236, 255],
[233, 235, 236, 255],
...,
[205, 206, 203, 255],
[204, 206, 203, 255],
[203, 206, 203, 255]],
[[233, 235, 236, 255],
[233, 235, 236, 255],
[233, 235, 236, 255],
...,
[205, 206, 203, 255],
[205, 206, 203, 255],
[204, 206, 203, 255]],
[[233, 235, 236, 255],
[233, 235, 236, 255],
[233, 235, 236, 255],
...,
[205, 206, 203, 255],
[205, 206, 203, 255],
[205, 206, 203, 255]]], dtype=uint8)